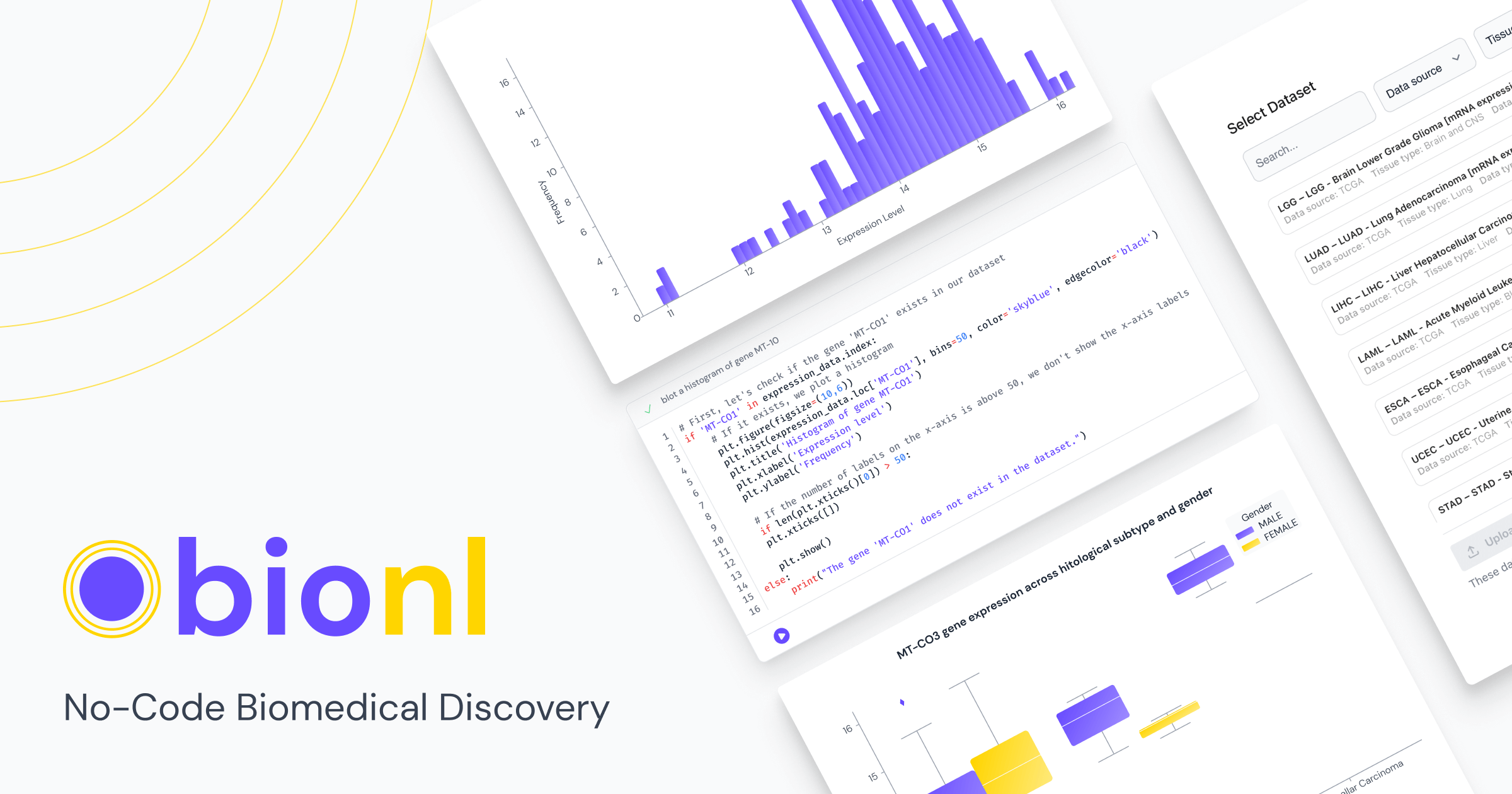
Navigating the Genome: From Sequencing to Cancer Insights

Introduction
In the evolving landscape of cancer research, high-throughput sequencing technologies have become a beacon, extracting vast raw genomic data. This wealth of information, however, is merely a starting point. The true challenge—and opportunity—lies in refining this data into actionable insights, aiming to increase our knowledge for genomic data, holding our life secrets.
Demystifying the Genetic Puzzle
The process begins as sequencers generate millions of DNA snippets. Bioinformatics algorithms could map these fragments against reference genomes, reconstructing the patient's unique genetic blueprint with unparalleled precision.
Sequencing techniques could be categorized based on their function and output:
1. By Read Length:
- Short-read sequencing
- Long-read sequencing
2. By Technology:
- Sanger sequencing: The traditional method, it uses chain termination to determine DNA sequence one base at a time. Highly accurate but slow and expensive.
- Next-generation sequencing (NGS): A broad term encompassing various high-throughput technologies like Illumina and Ion Torrent. Faster and cheaper than Sanger but with potential for errors.
The following Thermo Fisher Scientific video give a brief on how to determine what technology of sequencing to use, Sanger or NGS.
- Single-molecule real-time (SMRT) sequencing: It reads DNA sequences as they pass through a nanopore in real-time. Offers long reads but lower accuracy than other platforms. Learn more from this PacBio video.
- Nanopore sequencing: Employed by Oxford Nanopore Technologies (ONT), it directly reads electrical signals generated as DNA strands pass through a nanopore. Provides long reads with potential for lower cost compared to PacBio. ONTs video illustrates the sequencing process.
The choice of technique depends on various factors like research aims, budget, desired read length, and accuracy requirements. It's crucial to consider these factors carefully to select the most suitable technique for your specific research needs.
3. By Application:
- Whole-genome sequencing (WGS): Sequences the entire genome of an organism, used for studying genetic variation and identifying disease-associated mutations.
- Whole-exome sequencing (WES): Focuses on the protein-coding regions of genes, offering a cost-effective alternative to WGS for targeted variant detection.
- Targeted sequencing: Zooms in on specific genes or regions of interest, ideal for confirming known mutations or analyzing specific pathways.
- RNA sequencing (RNA-seq): Measures gene expression levels by sequencing RNA molecules, providing insights into cellular processes and disease mechanisms.
Transforming Data into Clinical Insight
The culmination of sequencing journey is a mosaic of genomic insights, highlighting potential biomarkers, therapeutic targets, and avenues for personalized medicine. These insights empower healthcare professionals to tailor treatment strategies to individual patients, ushering in a new era of precision oncology.
Bionl will help you overcome challenges:
- Navigating Complexity: Bionl's user friendly platform will provide you with the tools to analyze your data easily, no coding experience required.
- Ensuring Privacy and Security: At Bionl, data integrity and security are paramount. Our platform adheres to stringent ethical standards, ensuring that patient data is protected at every step.
- Fostering Collaboration: By standardizing data formats and analysis protocols, Bionl promotes global collaboration, accelerating the pace of discovery and innovation in cancer research.
Looking to the Future
As sequencing technologies advance and Bionl's analytical capabilities grow, the journey from raw data to revelatory insights will become ever more efficient. This progress promises not only deeper understanding of genomics data, but also more effective treatments, earlier diagnoses, and, ultimately, enhanced patient outcomes. By deciphering the complexities of cancer genomics, Bionl is not just contributing to the battle against cancer; it is redefining it, paving the way for a future where cancer can be comprehensively understood, effectively treated, and perhaps one day, overcome.